Exploring variant effects to advance precision medicine with Wellcome Connecting Science
A synopsis by Dr Sounak Sahu
Dr Sounak Sahu is a postdoctoral fellow at the National Cancer Institute (NCI), National Institutes of Health (NIH), USA and his research focuses on the development of Multiplexed Assays for Variant effects (MAVE) for breast cancer susceptibility genes. His long-term goal is to generate a sequence-function map for the entire BRCA2 gene, and other breast cancer susceptibility genes, which will provide a direct interpretation and proactive evidence for the effect of variants yet to be identified in the population.
Having recently attended the Curating the Clinical Genome, and the Mutational Scanning Symposium (both organised by the Wellcome Connecting Science (WCS) Learning and Training team), Dr Sahu shares an excellent synopsis below of the research discussed at each event; emphasising the opportunities sought through five days of interdisciplinary knowledge exchange and networking.

I felt privileged to attend these events, held at the Hinxton Hall Conference Centre, on the Wellcome Genome Campus from 10-14th July 2023. They provided an unparalleled platform to present my research, engage with peers, and collaborate with experts from diverse fields.
I am deeply grateful to the Connecting Science bursary, and travel grant from NCI, which helped me to participate at these global conferences.
Dr Sounak Sahu, Postdoctoral Fellow, National Cancer Institute (NCI), National Institutes of Health, USA
Deeper insight into variant interpretation: Curating the Clinical Genome (CCG)
The CCG conference focused on the latest advancements and challenges in deep-mutational scanning, and how researchers and clinicians can leverage the expanding genomics data to enhance clinical decision-making and advance personalised patient care. The conference commenced with a keynote address by Dr Ambroise Wonkam, from John Hopkins University, USA. Dr Wonkam emphasised the importance of understanding African genomes in identifying disease drug targets, as all modern humans trace their ancestry to Africans; citing the presence of 10% more DNA in the African pan-genome compared to the current human genome. Dr Wonkam concluded his talk with a call for the “3MAG project”, which aims to sequence at least three million African genomes. This resonated strongly with the community; promoting encouragement for equitable, innovative, and sustainable genetic medicine.
There’s 10% more DNA in the African pan-genome compared to the current human genome!
Dr Ambroise Wonkam, Johns Hopkins University, USA
The subsequent session explored genetic locus heterogeneity, genetic pleiotrophy, coding and non-coding genome variations, and the development of new methodologies to improve established functional assays and enhance variant classification. Notably, talks by Dr Nadia Carstens, on genomic medicine in Africa, and Dr Elizabeth McNally, on the monogenic and polygenic risk of heart disease, shed light on the importance of exome sequencing for rare developmental disorders and genetic variation of cardiomyopathies. Furthermore, discussions led by Dr Heide Cope emphasised the challenge of integrating genomic findings into clinical practice, and the role of ClinGen in generating guidelines for variant interpretation. Dr Andrew Wilkie provided valuable insights into the pathogenic variants in FGFR1 and FGFR3 genes associated with craniosynostosis. Dr Wilkie also proposed an innovative approach using patient-derived iPSCs, to complement and potentially replace, the resource-intensive mouse models.
The second day of CCG began with a captivating presentation by Dr Segun Fatumo, reiterating the significance of the African Genome and the vast genetic variations present within Africans compared to Eurasians. He highlighted the growing availability of large-scale African genome resources, emphasising their critical role in achieving genetic risk prediction. The subsequent sessions focused on clinical variant classification, and how non-coding regions are equally important for variant classification. Dr Nicky Whiffin emphasised the relevance of non-coding regions, which constitute a substantial portion of the genome, and highlighted the need for epigenetic data, as well as silico tools, to accurately interpret genetic variants. This session was followed by Dr Leslie Biesecker putting forward the recommendations on the interpretation of variants, outlining improvements from past guidelines. His informative talk covered the historical development of variant classification recommendations from 2000 to the most recent version in 2023. Dr Biesecker provided essential insights into defining variant “classification” and “interpretation”, encouraging a standardised approach in the field. Moreover, he addressed the importance of designating changes as “variants” rather than “mutation” or “polymorphisms”.
The final session of CCG focused on the functional assays, where I had the privilege to present our research on Saturation genome editing (SGE) of BRCA2 variants, along with Sofia Obolenski talking about the classification of POT1 variants, and Dr Haico van Attikum discussing PALB2 missense variants, shedding light on their implications in breast cancer susceptibility.
Clinical Application of MAVE Data workshop

The conclusion of CCG – with its strong emphasis on functional assays – provided a seamless transition to the MAVE workshop, which served as a perfect blend of ideas, discussing the utilisation of MAVE data for clinical variant classification. Dr Alan Rubin highlighted the curation of MAVE-DB website, an open access resource that allows sharing variant data from multiple genes obtained through orthogonal assays. This collaboration, and data sharing initiative, foster the advancement of variant classification. Dr Greg Findlay, one of the pioneers of SGE, shared valuable insights into the power and experimental strategies required for successful SGE experiments.
The MAVE workshop provided an enlightening learning experience for all attendees and the synergy between functional assays and data sharing initiatives showcased at the workshop is a testament to the collaborative spirit of the scientific community in the pursuit of precision medicine.
Dr Sounak Sahu, Postdoctoral Fellow, National Cancer Institute (NCI), National Institutes of Health, USA
Understanding the landscape of deep mutational scanning
The workshop was the perfect introduction to the Mutational Scanning Symposium (MSS), jointly organised by the WCS Learning and Training team and the Atlas of Variant Effect (AVE) alliance. The keynote address by Dr Fritz Roth described the genotype to phenotype problem, and the impact of environment in this relationship. The power of deep mutational scanning was demonstrated through variant atlas maps for 15-18 proteins with a detailed discussion on HMBS, MTHFR and LDLR genes.
Sketch of Fritz Roth's talk on 'Working towards a contextual atlas of variant effects'. Image credit: Alex Cagan
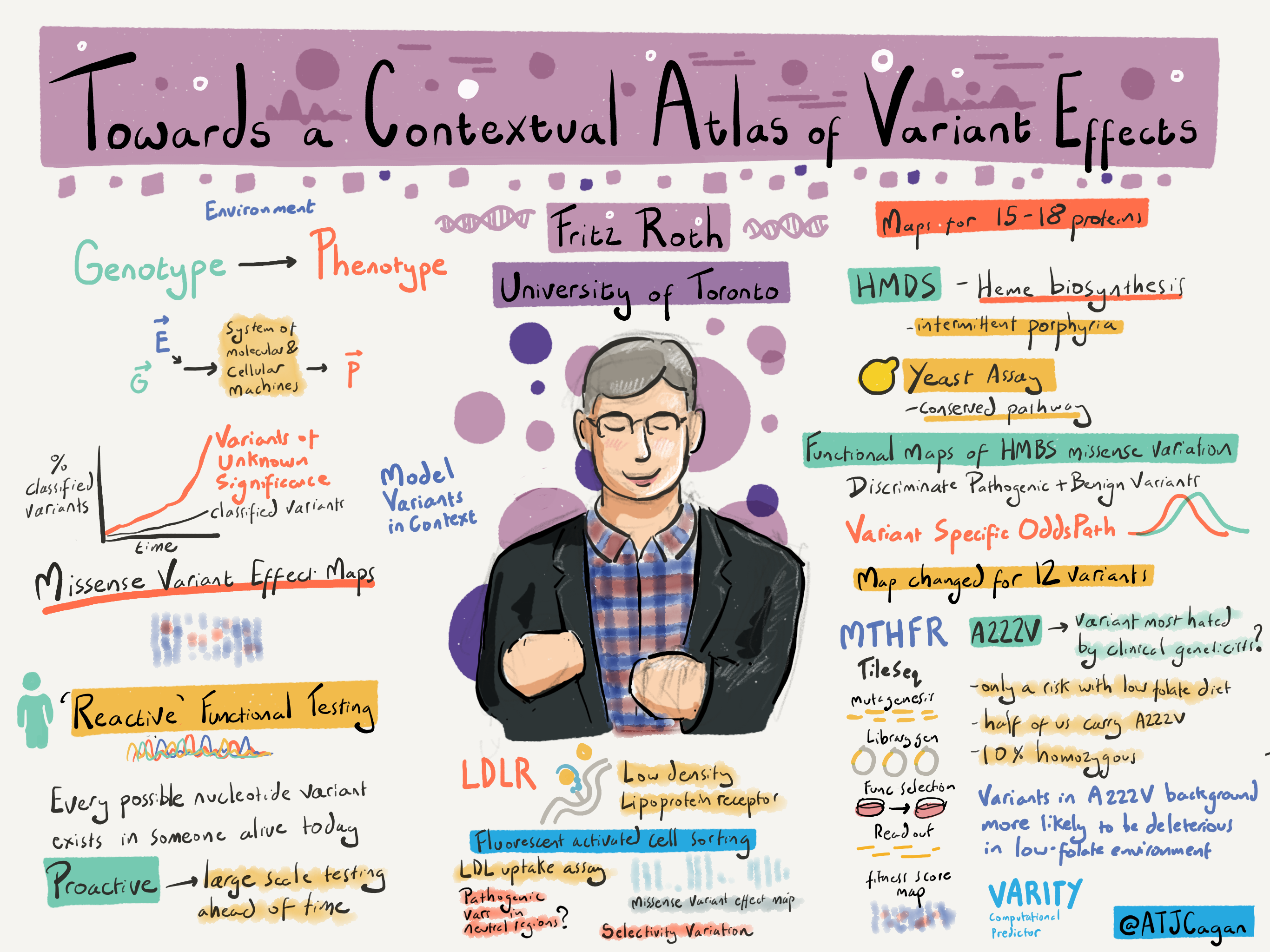
This was followed by others in the field discussing several other genes to develop MAVE approaches using SGE and cellular essentiality genetic screens. Talks on various genes, including BRCA1, VHL, NF2, JAG1 and others, showcased how mutational scanning facilitates genetic risk prediction. Dr Hong Kee Tan described the ongoing efforts by the Wellcome Sanger Institute for large-scale CRISPR screening for 200 genes in the next few years. The symposium also highlighted the importance of computational analysis, as Dr Mafilda Dias discussed the EVE score utilising evolutionary constraint and human population data.
The final day, I had the opportunity to chair the session on the development of new experimental tools using CRISPR, Dr Liselot Dewachter talking about genome editing in E. coli, Basheer Becerra on saturation mutagenesis screen to infer genotype-to-phenotype relationship. Dr Lea Starita showed a novel approach to utilise prime editing for variant classification by sequencing the pegRNAs harboring missense variants. Moreover, innovative applications of mutational scanning were demonstrated, such as thermostable vaccine development for SARS-CoV2 and rapid epitope mapping.
It was a wonderful experience learning about the novel technologies and hearing about unpublished work. As I return to my research at NCI, I am more inspired than ever to contribute to this rapidly evolving landscape of genomics and precision medicine.
Dr Sounak Sahu, Postdoctoral Fellow, National Cancer Institute (NCI), National Institutes of Health, USA
After this experience, I strongly believe that the combined effort of this scientific community and the Atlas of Variant Effect (AVE) alliance, alongside continuous advancements in genomics and data curation, will enable targeted therapies and personalised medicine to be realised. It’s more attainable than ever before!
For more insights from the Mutational Scanning Symposium, click here to read another blog article from our colleagues at the Wellcome Sanger Institute.
