
Advanced Courses and Scientific Conferences in partnership with FutureLearn launch a third online course – Bacterial Genomes: Accessing and Analysing Microbial Genome Data
Introducing Artemis - Bridging the gap between sequenced data and biological understanding
For years researchers have been trying to accurately interpret the biological significance of all their sequenced data in a meaningful manner, building on current understanding and furthering research.
With initiatives such as the 100K Pathogen Genome Project there is a wealth of large bacterial genome data sets being produced. . But their real biological value relies on researchers being able to visualise, browse and annotate this data effectively, using high-quality tools.
One of these genome browser and annotation tools, written specifically for the interrogation of smaller pathogens, archaea and lower eukaryotes, is Artemis; aptly named after the Greek goddess of the hunt.
What makes Artemis so powerful?
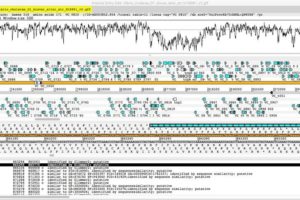
For scientists involved in the genome sequencing of small bacterial genomes it’s often the quality of the curated data that determines its success in helping the community understand more about disease-causing bacteria and how to track them. Raw sequence data offers little value to wider scientific understanding if it isn’t properly analysed using appropriate annotation tools to extract its biological importance.
Artemis is an annotation tool developed by researchers at the Wellcome Sanger Institute, as well as an active community of laboratory biologists and genome curators, built specifically around the needs of scientists, and with smaller (pathogen) genomes in mind.
One of these leading Sanger Institute researchers is Dr Julian Parkhill, Head of infection Genomics and a Senior Group Leader. Using Artemis during its early developmental phase to annotate Campylobacter jejuni, he was able to provide live scenario feedback to optimise the completion of this software, pushing its boundaries and actively cultivating a tool responsive to the community it was being designed for.
You can hear from Professor Julian Parkhill about the evolution of Artemis by enrolling on our forthcoming online course – Bacterial Genomes: Accessing and Analysing Genome Data, and listening to the interviews with our leading researchers.
Its focus on small pathogen genomes, with a six frame translation representation ability, enables it to interrogate and visualise this kind of data to a greater level of comparative analysis than is often possible with alternative genome browsers. It’s this that gives Artemis its versatility and makes it the annotation tool of choice for many scientists involved in the exploration of bacterial pathogen genomes.
“We use Artemis to visualise outputs from TraDIS experiments (a mutagenesis-based technique that allows you to very quickly profile the effects of different mutations on all of the genes in the genome at the same time). Artemis is fantastic because it allows you to follow up in a really high level of detail, because you can see the location of every mutation in your experience before and after infection.” – Dr Francesca Short (Scientist, Wellcome Sanger Institute)
Where can you learn more about the scientific value of Artemis?
Artemis is more than just an annotation tool – it enables scientists to bring their work to life and share new understanding visually.
Discovering how a tool like Artemis really works is the key to demonstrating how microbial bioinformatics delivers value in the real-world, and how sequenced data is taken from a raw format and interpreted.
Advanced Courses and Scientific Conferences, in partnership with FutureLearn the digital and social learning experts, will be launching a third online course, Bacterial Genomes: Accessing and Analysing Microbial Genome Data on 17 September, 2018. This internationally accessible, free online course will provide a detailed understanding of how genomic data and computational tools can help us to understand and track disease-causing bacteria, contributing to diagnosis and healthcare options. The course will demonstrate the analytical power of Artemis when exploring the genomic features of pathogens, and investigating whole bacterial genomes, as well as specific genes and proteins.
Sponsored by Advanced Courses and Scientific Conferences, the course is offered with full access completely free, including assessment and certification.
This course is designed for anyone with an interest in learning how to use tools to investigate bacterial genomes, and acquire bioinformatics skills to evaluate the role of microbial genes in disease.
The course will also be of interest to undergraduates, post-graduates, researchers, bioinformaticians, microbiologists, and healthcare professionals. In addition, the course is a powerful learning tool for Higher Education teachers looking for innovative methods to introduce genomic annotation and genome browsers to their students. Ideal for inclusive in class reading lists, or integration into lesson plans or lecture programmes.
Enrol today: http://bit.ly/2LQoWWx
Follow us on Twitter: @ACSCevents #FLaccess2analysis
